Final Project Idea
BioPainting with DNA - High resolution nanometric images
Goal: Create a stable 1M 2D image, with pixel size 10nm
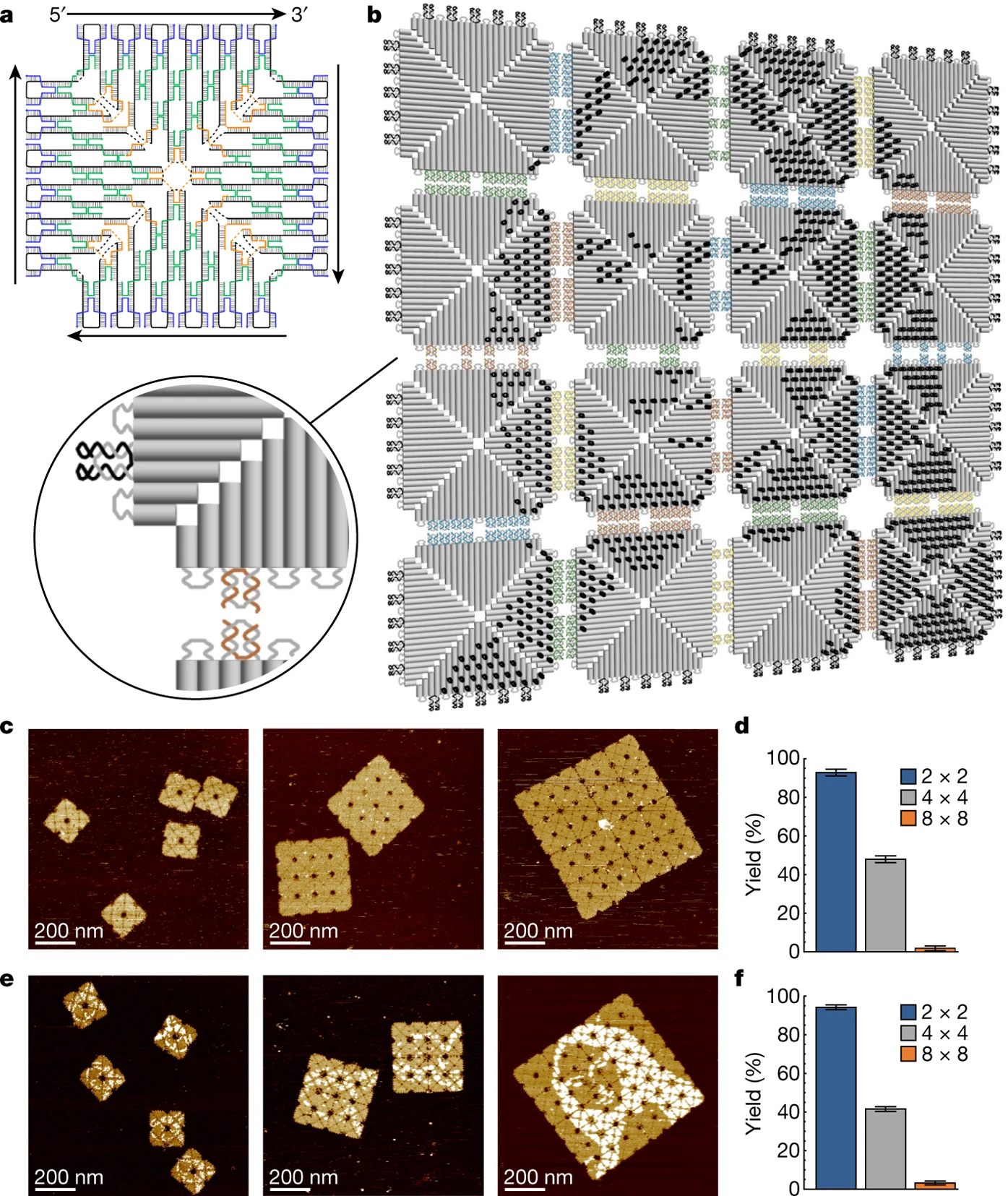
Prior Art: Fractal assembly of micrometre-scale DNA origami arrays with arbitrary patterns (Tikhomirov 2017)
Tikhomirov et al. used DNA origami tiles that are designed with specific “pixels” along each tile. Those tiles self assemble in a unique singular way to create nanometric images. Pixel size is approx. 6nm and the largest images contain ~9000 pixels. The images are not totally smooth due to DNA origami tile constraints.
Idea: Use DNA Microscopy by iterative ligation to create larger, smoother images.
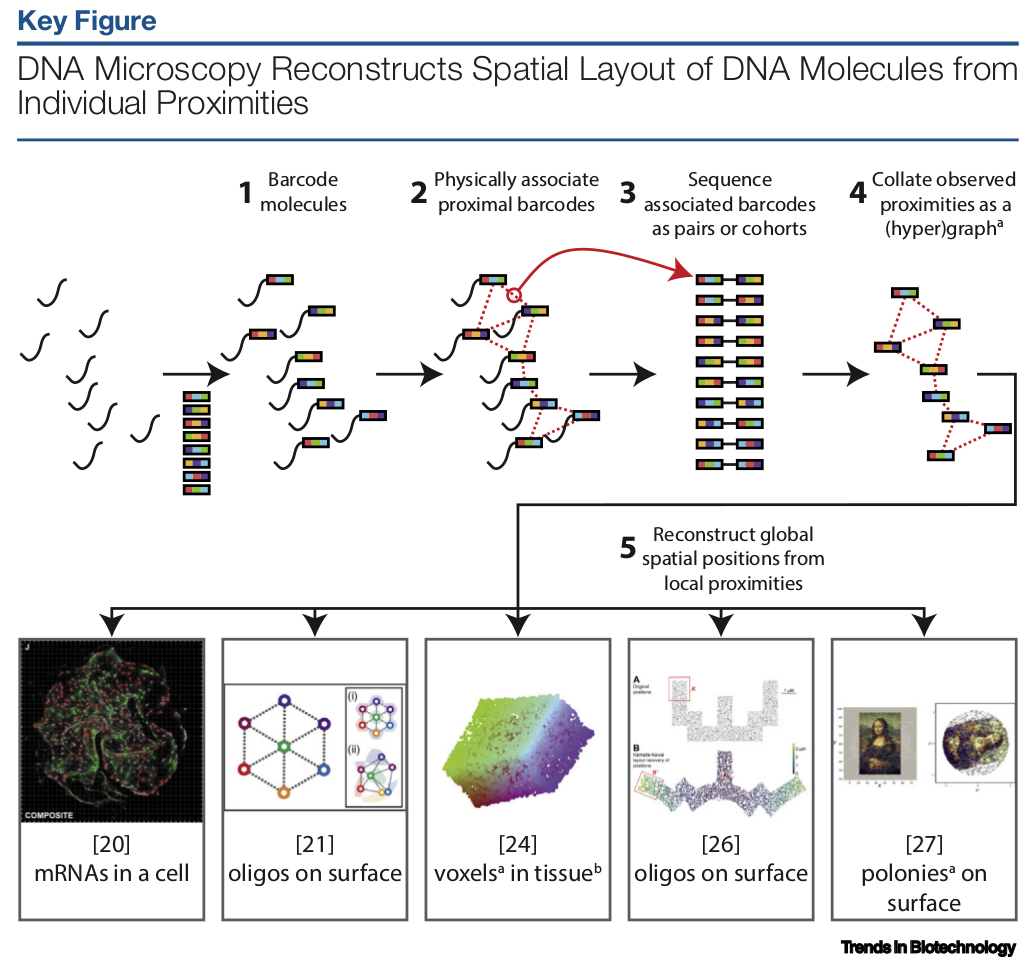
DNA microscopy is a novel technique done by multiple groups around the world. The basic concept is as follow:
- Generate single stranded DNA with unique random barcodes
- Immobilize barcodes to a surface of interest
- Apply a protocol where each neighboring strands are connected by a connector strand, ligated by DNA ligase, polymerized by DNA polymer. The extra strand is then melt off and the process repeats.
- At the end of the protocol, you have a tube with short DNA strands that contains pairs of barcodes. Each pair correspond to barcodes which are in proximity on the surface.
- By sequencing and a reconstrcution algorithm, it’s possible to attach a 2D coordinate to each random barcode.
 (Bringing Microscopy-By-Sequencing into View, Boulgakov 2019)
(Bringing Microscopy-By-Sequencing into View, Boulgakov 2019)
The above method can be used for various purposes, such as mapping connectomes, cellular mRNA, voxels in tissues and more…
What I am interested in is to use the same method for synthetic biology. While other methods focus on in vivo, what would could we accomplish if we use it in vitro?
We will create a chip that contains these barcodes. Then following the protocol, we would have a 2D coordinate and DNA barcode for each spot on the chip. These spots resolution are dependent on a few things:
- Streptavidin size (6nm) and number of binding sites
- DNA probe length
- Sequencing capacity
- Reconstruction algorithm accuracy
The number of barcodes can be extremely large.
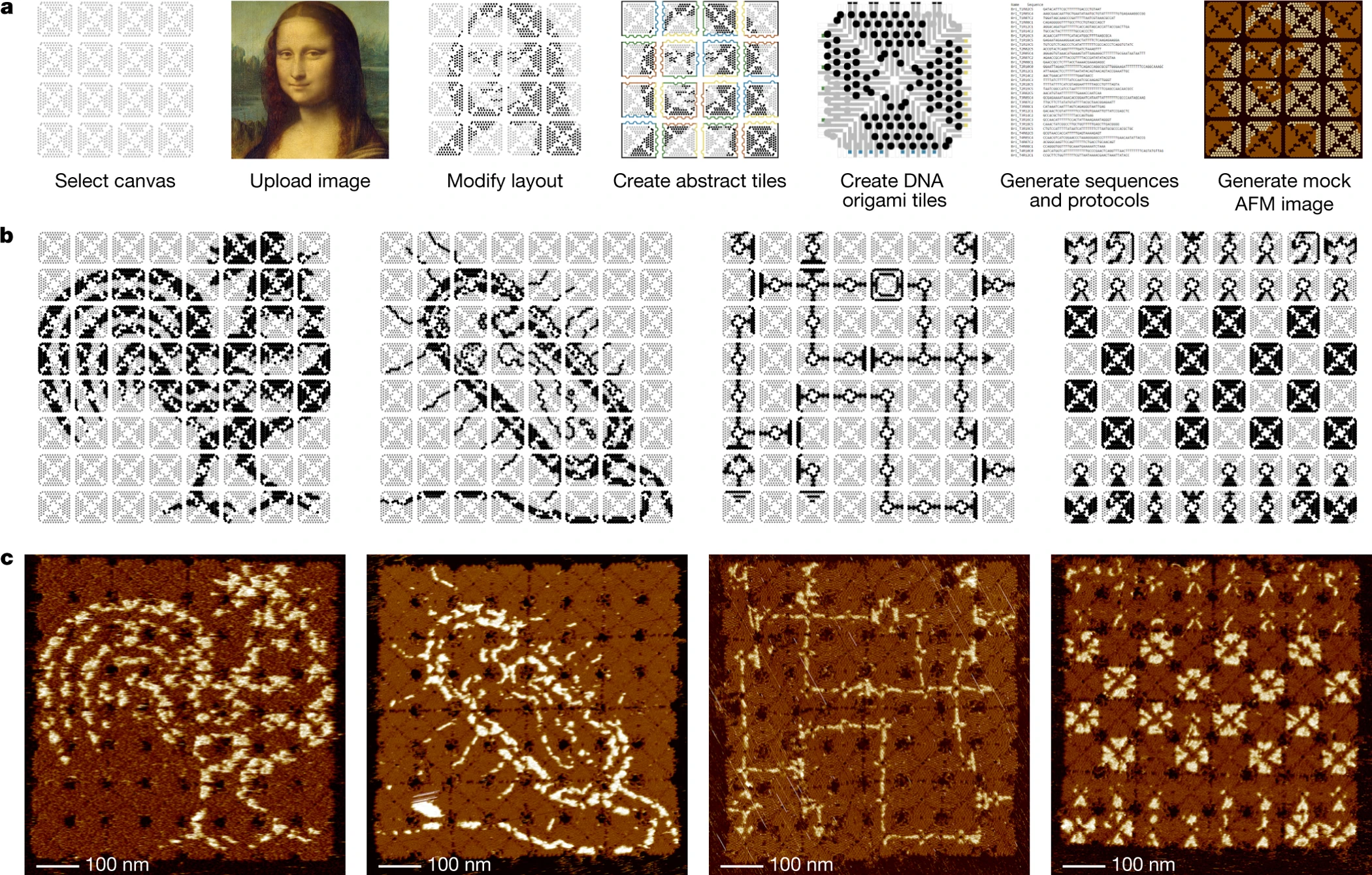
So what can we do with this barcoded chip? The simplest Proof of Concept would be to paint it, turn specific addresses ON and OFF, and create black & white images such as the Mona Lisa. If this proof of concept works, it could unlock a path towards more applications. We don’t have to stick to black and white, and go to full color. For more practical applications, there has been similar projects that use DNA origami to immobilize proteins as computational gates on a surface and create circuits.
For this class, I’m going to prepare a grant proposal with details on how to create this proof of concept.
The first steps would be to recreate the DNA microscopy protocol and characterize it, how many Streptavidin molecules bound to a specific area? How many DNA probes bind to each Streptavidin? How many DNA barcodes can we reconstruct and in which accuracy?
Next, the novel approach of addressing each of these barcodes after spatially locating them. We need to consider various methods such as DNA chips, CRISPR pooled libraries or more in order to generate a library of unique 1M barcodes, each with a “pixel color”.